Functions | |
| h5_err_t | H5PartSetNumParticles (h5_file_t *f, h5_size_t nparticles) |
| h5_err_t | H5PartSetNumParticlesStrided (h5_file_t *f, h5_size_t nparticles, h5_size_t stride) |
| h5_err_t | H5PartSetChunk (h5_file_t *f, h5_size_t size) |
| h5_ssize_t | H5PartGetNumDatasets (h5_file_t *f) |
| h5_err_t | H5PartGetDatasetName (h5_file_t *f, const h5_id_t idx, char *name, const h5_size_t len) |
| h5_err_t | H5PartGetDatasetInfo (h5_file_t *f, const h5_id_t idx, char *dataset_name, const h5_size_t len_dataset_name, h5_int64_t *type, h5_size_t *nelem) |
| h5_ssize_t | H5PartGetNumParticles (h5_file_t *f) |
| h5_err_t | H5PartResetView (h5_file_t *f) |
| h5_err_t | H5PartHasView (h5_file_t *f) |
| h5_err_t | H5PartSetView (h5_file_t *f, h5_int64_t start, h5_int64_t end) |
| h5_err_t | H5PartSetViewIndices (h5_file_t *f, const h5_id_t *indices, h5_size_t nelems) |
| h5_err_t | H5PartGetView (h5_file_t *f, h5_int64_t *start, h5_int64_t *end) |
| h5_err_t | H5PartSetCanonicalView (h5_file_t *f) |
Function Documentation
| h5_err_t H5PartSetNumParticles | ( | h5_file_t * | f, |
| h5_size_t | nparticles | ||
| ) |
Set the number of particles for the current time-step. After you call this subroutine, all subsequent operations will assume this number of particles will be written.
For the parallel library, the nparticles value is the number of particles that the individual task will write. You can use a different value on different tasks. This function uses an MPI_Allgather call to aggregate each tasks number of particles and determine the appropiate offsets. Because of the use of this MPI collective, it is advisable to call this function as few times as possible when running at large concurrency.
This function assumes that your particles' data fields are in stored in contiguous 1D arrays. For instance, the fields x and y for your particles are stored in separate arrays x[] and y[].
If instead you store your particles as tuples, so that the values are arranged 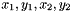 ... than you need to setup striding (in this case with value 2) using H5PartSetNumParticlesStrided.
... than you need to setup striding (in this case with value 2) using H5PartSetNumParticlesStrided.
- Returns:
H5_SUCCESSor error code
- Parameters:
-
[in] f Handle to open file [in] nparticles Number of particles
| h5_err_t H5PartSetNumParticlesStrided | ( | h5_file_t * | f, |
| h5_size_t | nparticles, | ||
| h5_size_t | stride | ||
| ) |
Set the number of particles for the current time-step. After you call this subroutine, all subsequent operations will assume this number of particles will be written.
For the parallel library, the nparticles value is the number of particles that the individual task will write. You can use a different value on different tasks. This function uses an MPI_Allgather call to aggregate each tasks number of particles and determine the appropiate offsets. Because of the use of this MPI collective, it is advisable to call this function as few times as possible when running at large concurrency.
This function assumes that your particles' data fields are stored tuples. For instance, the fields x and y of your particles are arranged 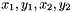 ... in a single data array. In this example, the stride value would be 2.
... in a single data array. In this example, the stride value would be 2.
If you instead have a separate array for each fields, such as x[] and y[], use H5PartSetNumParticles.
- Returns:
H5_SUCCESSor error code
- Parameters:
-
[in] f Handle to open file [in] nparticles Number of particles [in] stride Stride value (e.g. number of fields in the particle array)
| h5_err_t H5PartSetChunk | ( | h5_file_t * | f, |
| h5_size_t | size | ||
| ) |
Define the chunk size and enables chunking in the underlying HDF5 layer.
Note that this policy wastes disk space, but can improve write bandwidth on parallel filesystems that are sensitive to write alignment (e.g. lustre). It is only recommended when using the MPI-POSIX or MPI-IO independent VFDs (see H5OpenFile).
- Returns:
H5_SUCCESSor error code
| h5_ssize_t H5PartGetNumDatasets | ( | h5_file_t * | f ) |
Get the number of datasets that are stored at the current time-step.
- Returns:
- number of datasets in current timestep or error code
- Parameters:
-
[in] f Handle to open file
| h5_err_t H5PartGetDatasetName | ( | h5_file_t * | f, |
| const h5_id_t | idx, | ||
| char * | name, | ||
| const h5_size_t | len | ||
| ) |
This reads the name of a dataset specified by it's index in the current time-step.
If the number of datasets is n, the range of _index is 0 to n-1.
- Returns:
H5_SUCCESS
- Parameters:
-
[in] f Handle to open file [in] idx Index of the dataset [out] name Name of dataset [in] len Size of buffer name
| h5_err_t H5PartGetDatasetInfo | ( | h5_file_t * | f, |
| const h5_id_t | idx, | ||
| char * | dataset_name, | ||
| const h5_size_t | len_dataset_name, | ||
| h5_int64_t * | type, | ||
| h5_size_t * | nelem | ||
| ) |
Gets the name, type and number of elements of a dataset based on its index in the current timestep.
Type is one of the following values:
H5_FLOAT64_T(forh5_float64_t)H5_FLOAT32_T(forh5_float32_t)H5_INT64_T(forh5_int64_t)H5_INT32_T(forh5_int32_t)
- Returns:
H5_SUCCESS
- Parameters:
-
[in] f Handle to open file [in] idx Index of the dataset [out] dataset_name Name of dataset [in] len_dataset_name Size of buffer dataset_name[out] type Type of data in dataset [out] nelem Number of elements.
| h5_ssize_t H5PartGetNumParticles | ( | h5_file_t * | f ) |
This function returns the number of particles in this processor's view, if a view has been set.
If not, it returns the total number of particles across all processors from the last H5PartSetNumParticles call.
If you have neither set the number of particles nor set a view, then this returns the total number of particles in the first data set of the current time step. Note that H5Part assumes that all data sets within a given time step have the same number of particles (although the number particles can vary across time steps).
If none of these conditions are met, an error is thrown.
- Returns:
- number of particles in current timestep or an error code.
- Parameters:
-
[in] f Handle to open file
| h5_err_t H5PartResetView | ( | h5_file_t * | f ) |
Reset the view.
- Returns:
H5_SUCCESS
- Parameters:
-
[in] f Handle to open file
| h5_err_t H5PartHasView | ( | h5_file_t * | f ) |
Check whether a view has been set, either automatically with H5PartSetNumParticles or manually with H5PartSetView or H5PartSetViewIndices.
- Returns:
- 0 for false or 1 for true
- Parameters:
-
[in] f Handle to open file
| h5_err_t H5PartSetView | ( | h5_file_t * | f, |
| h5_int64_t | start, | ||
| h5_int64_t | end | ||
| ) |
For parallel I/O or for subsetting operations on the datafile, this function allows you to define a subset of the total particle dataset to operate on. The concept of "view" works for both serial and for parallel I/O. The "view" will remain in effect until a new view is set, or the number of particles in a dataset changes, or the view is "unset" by calling H5PartSetView(file,-1,-1);
Before you set a view, H5PartGetNumParticles will return the total number of particles in the current time-step (even for the parallel reads). However, after you set a view, it will return the number of particles contained in the view.
The range is inclusive: the end value is the last index of the data.
- Returns:
H5_SUCCESSor error code
- Parameters:
-
[in] f Handle to open file [in] start Start particle [in] end End particle
| h5_err_t H5PartSetViewIndices | ( | h5_file_t * | f, |
| const h5_id_t * | indices, | ||
| h5_size_t | nelems | ||
| ) |
For parallel I/O or for subsetting operations on the datafile, this function allows you to define a subset of the total dataset to operate on by specifying a list of indices. The concept of "view" works for both serial and for parallel I/O. The "view" will remain in effect until a new view is set, or the number of particles in a dataset changes, or the view is "unset" by calling H5PartSetViewIndices(NULL,0);
Before you set a view, the H5PartGetNumParticles() will return the total number of particles in the current time-step (even for the parallel reads). However, after you set a view, it will return the number of particles contained in the view.
- Returns:
H5_SUCCESSor error code
- Parameters:
-
[in] f Handle to open file [in] indices List of indices [in] nelems Size of list
| h5_err_t H5PartGetView | ( | h5_file_t * | f, |
| h5_int64_t * | start, | ||
| h5_int64_t * | end | ||
| ) |
Allows you to query the current view. Start and End will be -1 if there is no current view established. Use H5PartHasView() to see if the view is smaller than the total dataset.
- Returns:
- number of elements in the view or error code
- Parameters:
-
[in] f Handle to open file [out] start Start particle [out] end End particle
| h5_err_t H5PartSetCanonicalView | ( | h5_file_t * | f ) |
If it is too tedious to manually set the start and end coordinates for a view, the H5SetCanonicalView() will automatically select an appropriate domain decomposition of the data arrays for the degree of parallelism and set the "view" accordingly.
- Returns:
- H5_SUCCESS or error code
- Parameters:
-
[in] f Handle to open file
 1.7.2
1.7.2